 |
Bimetric 3+1 toolkit for spherical symmetry
|
 |
Bimetric 3+1 toolkit for spherical symmetry
|
Time evolution using the Method of Lines (MoL). More...
#include <integrator.h>
Public Member Functions | |
| MoL (Parameters ¶ms, UniformGrid &ug, GridOutputWriter &out) | |
| Constructs the MoL integrator as specified in the parameter file. | |
| ~MoL () | |
| The destructor (here only responsible for cleaning up standard output). | |
| void | quit (Int m=-1) |
| Moves the integration final time to the current time and marks the integration stopped. | |
| void | addToEvolution (IntegFace *eomInterface) |
| Adds the EoM to the list of all evolved subsystems. | |
| void | keepConstant (const std::vector< Int > &gfs) |
| The given fields will be kept constant by the integrator. | |
| void | keepEvolved (const std::vector< fld::EvolvedBy > &gfs) |
| Add the given functions to the evolution list. | |
| Real | dt () const |
| Returns the time step. | |
| bool | evolveEquations () |
| Integrates the evolution equations in the given grid realm. | |
Private Member Functions | |
| void | reportIntegrationTime (Int m) |
| Report the integration time and the estimated real-time to finish. | |
| void | integStep_Begin (Int m) |
| Executed at the beginning of each integration step. | |
| void | integStep_End (Int m1, Int m) |
| Executed at the end of each integration step. | |
| void | integStep_Checkpoint (Int m) |
| Executed at each checkpoint. | |
| void | integStep_Euler (Int mNext, Int m) |
| Evolves the state variables using the FT scheme. | |
| void | integStep_AvgICN (Int mNext, Int mMid, Int m) |
| Evolves the sate variables using the ICN averaging. | |
| void | integrate_AvgICN (int ICN_steps) |
| Time evolution using the iterated Crank-Nicolson (ICN). | |
| void | integStep_MoLInit (Int m1, Int m, Real beta_delta_t) |
| Prepares the state variables for the intermediate MoL steps. | |
| void | integStep_MoL (Int m1, Int m, Real alpha) |
| The intermediate method of lines (MoL) steps. | |
| void | integrate_MoL (const MoLDescriptor &MoL) |
| Time evolution using the method of lines (MoL). | |
Static Private Member Functions | |
| static void | signalHandler (int signum) |
| Forward the signal to all integrators (signaling them to quit). | |
Private Attributes | |
| std::vector< IntegFace * > | eomList |
| Equations of motion that we evolve. | |
| std::vector< Int > | constantGF |
| Grid functions that are kept constant. | |
| std::vector< fld::EvolvedBy > | evolvedGF |
| Grid functions that are integrated in time. | |
| GridOutputWriter * | output |
| The integration output is directed here. | |
| time_hr | rt_start |
| The realtime when the integration started. | |
| Int | method |
| The integration method (Euler, ICN, MoL, ...) | |
| bool | running |
| Indicates if the integration should proceed. | |
| Real | t_0 |
The initial t. | |
| Real | t_1 |
Integrate up to this t. | |
| Real | delta_t |
| The integration step. | |
| Real | dissip |
| Kreiss-Oliger dissipation coefficient. | |
| Real | dissip_delta_r |
dissip / delta_r | |
| Real | cur_t |
| Current time. | |
Static Private Attributes | |
| static std::map< std::string, int > | knownMethods |
| A list of the implemented integration methods. | |
| static std::vector< MoL * > | knownIntegrators |
| Keep the list of all created integrator objects (used by signalHandler). | |
Time evolution using the Method of Lines (MoL).
The implemented methods are declared in MoL::knownMethods.
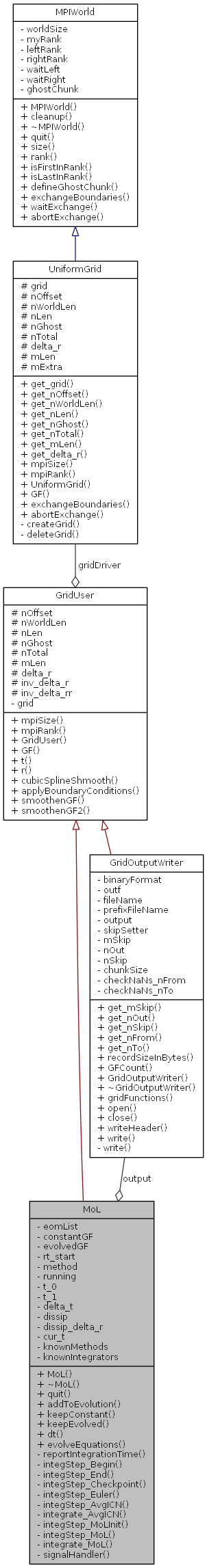
The integrator can evolve equations from several modules at the same time. All such modules should implement IntegFace ans sign in to the integrator. The modules should also specify which grid functions are either evolved or kept constant by the integrator. (Those grid functions that are unknown to the integrator have to be maintained by the the modules themselves.)
The system provides the coordinates t and r as global grid functions (they are defined in fld::sysIndex). The integrator keeps time t evolved and the spatial coordinate r constant.
The integrator handles process signals (e.g., Ctrl-C), which will cause a premature end of the integration.
Definition at line 31 of file integrator.h.
| MoL::MoL | ( | Parameters & | params, |
| UniformGrid & | ug, | ||
| GridOutputWriter & | out | ||
| ) | [inline] |
Constructs the MoL integrator as specified in the parameter file.
Definition at line 209 of file integrator.h.
: GridUser( ug ), output( &out ) { constantGF.reserve( 64 ); evolvedGF.reserve( 128 ); constantGF.push_back( fld::r ); // Radial coordinate is kept constant by default params.get( "integ.t_0", t_0, 0.0 ); params.get( "integ.t_1", t_1, 1.0 ); params.get( "integ.delta_t", delta_t, 0.1 ); params.get( "integ.dissip", dissip, 0.0 ); std::string name = params.get( "integ.method", method, 0, knownMethods ); // Fix the sign of delta_t // delta_t = t_1 < t_0 ? -abs(delta_t) : abs(delta_t); cur_t = t_0; // The initial `t` running = false; // Stopped integration by default (will be enabled later on) // Calculate Kreiss-Oliger dissipation corrected delta r // dissip_delta_r = dissip / delta_r; // Expected output file data size (in bytes) // const Real sz = ( ( t_1 - t_0 ) / delta_t / output->get_mSkip() + 1 ) * output->recordSizeInBytes(); slog << "Integrator:" << std::endl << std::endl << " t_0 = " << t_0 << ", t_1 = " << t_1 << ", delta_t = " << delta_t << ", method = " << name << " (#" << method << ")" << std::endl << " Kreiss-Oliger dissipation = " << dissip << std::endl << " Expected output data size = " << round( ( sz < 1e3 ? sz : sz < 1e6 ? sz/1e3 : sz/1e6 ) * 10 ) / 10 << ( sz < 1e3 ? "" : sz < 1e6 ? " kB" : " MB" ) << ", GF count = " << output->GFCount () << std::endl << std::endl; // Register to receive the signals from the system // knownIntegrators.push_back( this ); if ( knownIntegrators.size () == 1 ) { signal( SIGINT, signalHandler ); signal( SIGTERM, signalHandler ); } }
| MoL::~MoL | ( | ) | [inline] |
The destructor (here only responsible for cleaning up standard output).
Definition at line 261 of file integrator.h.
{
if( mpiRank() == 0 ) {
std::cout << std::endl << std::endl;
}
}
| void MoL::addToEvolution | ( | IntegFace * | eomInterface | ) | [inline] |
Adds the EoM to the list of all evolved subsystems.
Definition at line 288 of file integrator.h.
{
eomList.push_back( eomInterface );
}
| bool MoL::evolveEquations | ( | ) | [inline] |
Integrates the evolution equations in the given grid realm.
Definition at line 315 of file integrator.h.
{
#if 0
if( delta_r != r(0,nGhost+1) - r(0,nGhost) ) {
slog << " Error: delta_r = " << delta_r << " != grid spacing = "
<< r(0,nGhost+1) - r(0,nGhost)
<< ", diff = " << delta_r - ( r(0,nGhost+1) - r(0,nGhost) )
<< std::endl;
}
#endif
output->writeHeader ();
// Start timing the real-time
rt_start = std::chrono::high_resolution_clock::now ();
switch( method )
{
case 0: integrate_AvgICN ( 0 ); break;
case 1: integrate_AvgICN ( 1 ); break;
case 2: integrate_AvgICN ( 2 ); break;
case 3: integrate_AvgICN ( 3 ); break;
case 4: integrate_MoL ( ICN2 ); break;
case 5: integrate_MoL ( ICN3 ); break;
case 6: integrate_MoL ( RK1 ); break;
case 7: integrate_MoL ( RK2 ); break;
case 8: integrate_MoL ( RK3 ); break;
case 9: integrate_MoL ( RK4 ); break;
}
return true;
}
| void MoL::integStep_Begin | ( | Int | m | ) | [inline, private] |
Executed at the beginning of each integration step.
Definition at line 101 of file integrator.h.
{
/// - Prepare the intermediate variables.
///
for( auto eom: eomList ) {
eom->integStep_Prepare( m );
}
/// - Exchange boundaries with the neighboring MPI ranks and/or fix the
/// left/right boundary conditions (depending on our MPI rank).
/// Quit if MPI has failed or we received abort message.
///
if( gridDriver->mpiSize() > 1
&& ! gridDriver->exchangeBoundaries( m ) )
{
running = false;
return;
}
if ( gridDriver->isFirstInRank () ) {
for( auto eom: eomList ) {
eom->applyLeftBoundaryCondition( m );
}
}
if ( gridDriver->isLastInRank () ) {
for( auto eom: eomList ) {
eom->applyRightBoundaryCondition( m );
}
}
/// - Calculate the RHS of time evolution equations.
///
for( auto eom: eomList ) {
eom->integStep_CalcEvolutionRHS( m );
}
}
| void MoL::integStep_Checkpoint | ( | Int | m | ) | [inline, private] |
Executed at each checkpoint.
Definition at line 158 of file integrator.h.
{
/// - Report the elapsed (both integration and real) time.
///
reportIntegrationTime( m );
/// - Check for eventual NaNs in the central/lower region of the grid.
///
for( auto eom: eomList )
{
if ( ! eom->integStep_Diagnostics( m,
output->get_nFrom (), output->get_nTo () ) ) {
if ( running ) { // quit only once
quit( m );
}
}
}
/// - Send the grid slice to the output.
///
output->write( m, cur_t, delta_t );
}
| void MoL::integStep_End | ( | Int | m1, |
| Int | m | ||
| ) | [inline, private] |
Executed at the end of each integration step.
Returns false if we need to break the integration loop.
Definition at line 140 of file integrator.h.
{
/// - Finalize the integration step (e.g., doing diagnostic and post
/// processing of EoM like calculating the constraint violations).
///
for( auto eom: eomList ) {
eom->integStep_Finalize( m1, m );
}
/// - Wait on finishing the exchange of the boundaries (when using MPI).
///
if ( ! gridDriver->waitExchange () ) {
quit( m ); // Quit if MPI has failed
}
}
| void MoL::keepConstant | ( | const std::vector< Int > & | gfs | ) | [inline] |
The given fields will be kept constant by the integrator.
Definition at line 295 of file integrator.h.
{
constantGF.insert( constantGF.end(), gfs.begin(), gfs.end() );
}
| void MoL::keepEvolved | ( | const std::vector< fld::EvolvedBy > & | gfs | ) | [inline] |
Add the given functions to the evolution list.
Definition at line 302 of file integrator.h.
Moves the integration final time to the current time and marks the integration stopped.
Definition at line 271 of file integrator.h.
{
running = false; // Stop the integration
t_1 = m < 0 ? cur_t : GF( fld::t, m, nGhost );
if( mpiRank() == 0 ) std::cout << std::endl;
slog << "*** Quitting at t = " << t_1 << std::endl;
/// - In case of NaNs, signal the other MPI ranks that we are quitting.
/// @todo Distribute the abort msg to all ranks instead of calling MPI_Abort.
///
gridDriver->abortExchange ();
}
| void MoL::reportIntegrationTime | ( | Int | m | ) | [inline, private] |
Report the integration time and the estimated real-time to finish.
Definition at line 74 of file integrator.h.
{
if ( mpiRank() > 1 ) {
return; // only the first in rank can access the standard output
}
Real tnow = GF( fld::t, m, nGhost ); // current integration time
auto elapsed = 1e-3 * std::chrono::duration_cast<std::chrono::milliseconds>
( std::chrono::high_resolution_clock::now () - rt_start ).count ();
auto rt_factor = elapsed / ( tnow - t_0 ); // real time/integration time
auto rt_total = round( ( t_1 - t_0 ) * rt_factor ); // projected total time
auto rt_left = round( ( t_1 - tnow ) * rt_factor ); // estimated time to cover
std::cout << " t = " << std::setw(6) << std::setprecision(2) << std::fixed
<< tnow << std::setw(-1) << std::setprecision(-1) << std::defaultfloat
<< ", real = " << round( elapsed )
<< " s (" << round( 100 * ( tnow - t_0 ) / ( t_1 - t_0 ) )
<< "% of " << rt_total
<< " s), ETA " << rt_left << " s"
<< " \r";
std::flush( std::cout );
}
| static void MoL::signalHandler | ( | int | signum | ) | [inline, static, private] |
Forward the signal to all integrators (signaling them to quit).
Definition at line 59 of file integrator.h.
{
std::cout << std::endl;
std::cerr << "*** Signal " << signum << " received" << std::endl;
if( knownIntegrators.size () == 0 ) {
exit( -1 );
}
for( auto i: knownIntegrators ) {
i->quit ();
}
}
std::vector<Int> MoL::constantGF [private] |
Grid functions that are kept constant.
Definition at line 34 of file integrator.h.
Real MoL::cur_t [private] |
Current time.
Definition at line 47 of file integrator.h.
Real MoL::delta_t [private] |
The integration step.
Definition at line 44 of file integrator.h.
Real MoL::dissip [private] |
Kreiss-Oliger dissipation coefficient.
Definition at line 45 of file integrator.h.
Real MoL::dissip_delta_r [private] |
dissip / delta_r
Definition at line 46 of file integrator.h.
std::vector<IntegFace*> MoL::eomList [private] |
Equations of motion that we evolve.
Definition at line 33 of file integrator.h.
std::vector<fld::EvolvedBy> MoL::evolvedGF [private] |
Grid functions that are integrated in time.
Definition at line 35 of file integrator.h.
Int MoL::method [private] |
The integration method (Euler, ICN, MoL, ...)
Definition at line 40 of file integrator.h.
GridOutputWriter* MoL::output [private] |
The integration output is directed here.
Definition at line 37 of file integrator.h.
time_hr MoL::rt_start [private] |
The realtime when the integration started.
Definition at line 39 of file integrator.h.
bool MoL::running [private] |
Indicates if the integration should proceed.
Definition at line 41 of file integrator.h.
The initial t.
Definition at line 42 of file integrator.h.
Integrate up to this t.
Definition at line 43 of file integrator.h.